 AlphaFold neural network produced a ‘totally transformative’ database of more than 350,000 structures from Homo sapiens and 20 model organisms.
AlphaFold neural network produced a ‘totally transformative’ database of more than 350,000 structures from Homo sapiens and 20 model organisms.
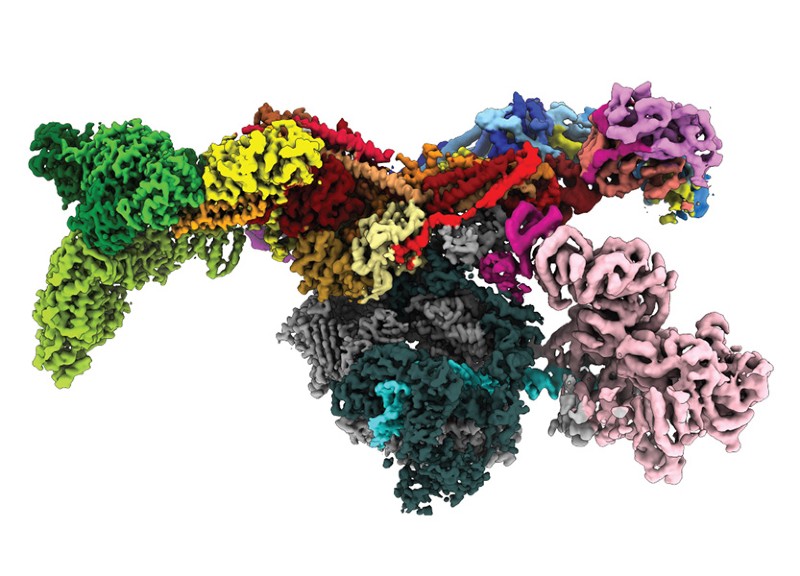
The human genome holds the instructions for more than 20,000 proteins. But only about one-third of those have had their 3D structures determined experimentally. And in many cases, those structures are only partially known.
Now, a transformative artificial intelligence (AI) tool called AlphaFold, which has been developed by Google’s sister company DeepMind in London, has predicted the structure of nearly the entire human proteome (the full complement of proteins expressed by an organism). In addition, the tool has predicted almost complete proteomes for various other organisms, ranging from mice and maize (corn) to the malaria parasite (see ‘Folding options’).
The more than 350,000 protein structures, which are available through a public database, vary in their accuracy. But researchers say the resource — which is set to grow to 130 million structures by the end of the year — has the potential to revolutionize the life sciences.























